Our Marie Curie Training Network “SmartNets” has been funded!
Scientists from five universities immerse themselves into the structure of networks. We will investigate how the highly advanced networks in biological systems, from molecules to the brain, are built and how information is transferred through them. The Marie Skłodowska Curie Innovative Training Network “SmartNets” has been funded at the end of 2019.
| more information: | SmartNets webstie |
Modelling sensory map plasticity in barrel cortex
An important feature of the brain is its ability to retain plasticity not only during development, but also later in life in response to experience. This experience dependent plasticity (EPD) modulates connections between neurons and is also present during sensory map reorganization. In barrel cortex, whisker deprivation (plucking a whisker) diminishes the input to its corresponding barrel, after which reorganization of the sensory maps takes place. This causes the representation of the deprived whisker to shrink, while the representation of surrounding whiskers expands. Koen Kole investigated how such sensory experience affects supragranular cross-columnar synaptic communication. Linda Wouters showed with a computational model which connectivity changes could explain these experimental results. Tousif Jamal is currently analyzing these data. Tea Tompos is investigating how sparsification of synaptic connectivity influences network homeostasis.
| link to code: | GitHub |
Computational model of a somatosensory cortical column
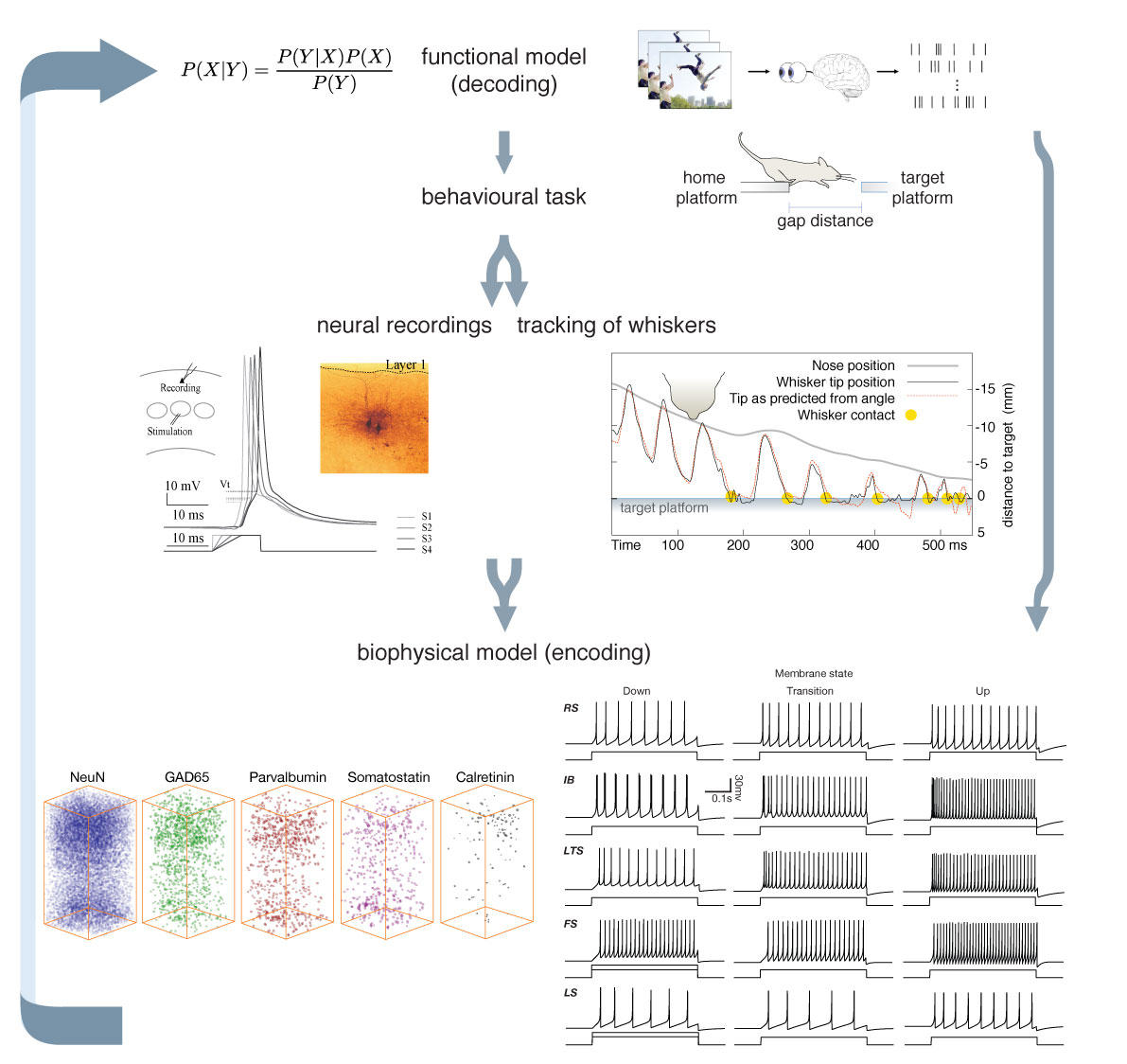
We continually change the way we perceive the world by actively changing the position and physical properties of our sensory organs, for instance by moving and focussing our eyes to pursue a moving object, yet we perceive objects in the world as stable entities. How the brain processes this changing sensory information, what computations it performs to create unvarying percepts and how the interaction between perception and active control of our sensory organs (sensorimotor integration) arises, is still largely unknown. I research these computations: How do neurons extract information about the world and represent it? How does the information sensory neurons extract influence active control over our sensory organs and vice versa?
Perception and sensorimotor integration are often studied in the rodent whisker system, for several reasons. Firstly, rodents use active sensorimotor integration to solve real-world problems like navigation, object localization and texture discrimination. Secondly, whisker motion can easily be tracked in millisecond resolution and manipulated experimentally, giving direct access to the sensory information that animals collect and to the motor plan they generate during touch sensing. Finally, the availability of molecular tools in the mouse makes it a good candidate to address neural circuit mechanisms. Even though the rodent whisker system has been experimentally well studied and lends itself to a mechanistic explanation of neural computation, there are not many computational models of it. Moreover, the models that have been proposed only describe certain modules in the whisker system, such as biomechanical whisker models, mechanoreceptor models, cortical models or behavioural models.
In this project, Chao Huang and me developed an end-to-end model to allow the integration of and cross-validation between structural and functional aspects of sensory phenomena. Nicolas Rault is currently defining models of brain stem and thalamus. We keep on updating the model with the latest insights. The model is based on anatomical reconstructions and includes known electrophysiological cell properties. The connectivity is probabilistic, so every time the model is run, a new realisation of the connectivity is established. The newest version can be found in the GitHub link below. Matteo Cantarelli of MetaCell LLC Ltd, and Salvador Dura-Bernal and Eugenio Urdapilleta of the Dura-Bernal Lab, SUNY Downstate have helped us to convert the model to NetPyNE.
| link to code: |
GitHub – biophysical network model |
| link to manuscript: | bioRxiv – network description, bioRxiv – information transfer and recovery |
| links to conference abstracts | CNS 2020 |
Balanced networks
Based on the network parameters measured for the computational model above, Prescilla Uijtewaal created a balanced network of L2/3 and L4 of the barrel cortex to see if the ‘balanced asynchronous state’ still holds for realistic numbers of neurons and connectivity. The network consists of integrate-and-fire neurons that are locally connected. These adaptations in the neural populations and connectivity result in different dynamic regimes. Additya Sharma and Ildefonso Ferreira Pica are investigating how the connectivity structure of the barrel cortex influences its information processing.
| link to code: | Github |
| links to conference abstracts | CNS 2020 |
FORCE learning for whiskers
To look at functional effects, Lino Vliex, Tim Winter, Lois de Groot and Hendrik Scheeres developed a trainable barrel cortical network in order to investigate whisker-related information processing. For this, we use a spike-based FORCE learning application. Hendrik Scheeres and Lois de Groot are/were extending this model to include dopaminergic modulation and Dale’s law.
| link to code: | Github – FORCE learning for whiskers |
| links to conference abstract | CNS 2020 |
Workshop on biological network analysis: 23 November 2018
Electrical activity in neural networks, transcriptions in genetic networks, the schooling of fish and diseases spreading in social networks, all biological networks share a common factor: the behaviour of a network is critically determined by its structure. This structure induces collective emergent behaviour that can only be understood by analysing the whole network in relation to its constituent parts.
The relation between network structure, network dynamics and information processing capacity is essential at every scale: from molecules and genes to large neural networks to populations of behaving agents: on every level nodes form complex networks underlying the most essential functions of the brain, body and society. Only recently, with high-throughput techniques, have we begun to collect the vast amounts of data needed to study the structure and functioning of such networks. However, analysing these data is still a challenge and the nature of complex network processes are still poorly understood. This workshop will bring together scientists studying biological networks across disciplines and dimensions, from molecule to independent agent and from data recording to network analysis and simulation.